Archives
- 2025-12
- 2025-11
- 2025-10
- 2025-09
- 2025-03
- 2025-02
- 2025-01
- 2024-12
- 2024-11
- 2024-10
- 2024-09
- 2024-08
- 2024-07
- 2024-06
- 2024-05
- 2024-04
- 2024-03
- 2024-02
- 2024-01
- 2023-12
- 2023-11
- 2023-10
- 2023-09
- 2023-08
- 2023-07
- 2023-06
- 2023-05
- 2023-04
- 2023-03
- 2023-02
- 2023-01
- 2022-12
- 2022-11
- 2022-10
- 2022-09
- 2022-08
- 2022-07
- 2022-06
- 2022-05
- 2022-04
- 2022-03
- 2022-02
- 2022-01
- 2021-12
- 2021-11
- 2021-10
- 2021-09
- 2021-08
- 2021-07
- 2021-06
- 2021-05
- 2021-04
- 2021-03
- 2021-02
- 2021-01
- 2020-12
- 2020-11
- 2020-10
- 2020-09
- 2020-08
- 2020-07
- 2020-06
- 2020-05
- 2020-04
- 2020-03
- 2020-02
- 2020-01
- 2019-12
- 2019-11
- 2019-10
- 2019-09
- 2019-08
- 2019-07
- 2019-06
- 2019-05
- 2019-04
- 2018-11
- 2018-10
- 2018-07
-
br Ubiquitin Capture by the Mobile
2020-03-24
Ubiquitin Capture by the Mobile APC11 RING Domain Contributes to Processive Substrate Ubiquitylation Although the rules of Ub-mediated proteolysis are only beginning to emerge, the rate and order in which different APC/C substrates are degraded during the Estradiol valerate correlates with proce
-
In addition to cleaving ubiquitins off modified proteins DUB
2020-03-24

In addition to cleaving ubiquitins off modified proteins, DUBs can also cleave between ubiquitin moieties within a polyubiquitin chain to edit the ubiquitin signal. DUBs employ different strategies to recognize polyubiquitin (Figure 1). DUBs that rely only on interactions on the S1 site tend to be C
-
In addition to glutamatergic inputs the RTn integrates
2020-03-24
In addition to glutamatergic inputs, the RTn integrates cholinergic, serotonergic and noradrenergic synapses of fibers coming from nuclei located in the brainstem (Asanuma, 1992, Morrison and Foote, 1986, Pare et al., 1988) involved in the alternating firing mode and in the frequency changes of reti
-
Employing distinct genetic and pharmacological approaches Di
2020-03-24
Employing distinct genetic and pharmacological approaches, Divakaruni et al. now demonstrate that macrophages still acquire their IL-4-elicited phenotype in the absence of CPT1a (considered the main isoform in leukocytes) or CPT2, and highlight that the increased LC-FAO in M(IL-4) cells is less cruc
-
PKA signalling in the nucleus
2020-03-24
PKA signalling in the nucleus was thought to be due to the translocation of the catalytic subunit upon activation from the 486 2 to the nucleus via diffusion [72]. However, a new understanding has emerged, as both the regulatory and catalytic subunits have been identified in the nucleus and functio
-
Introduction Quassinoids are natural products formed through
2020-03-24
Introduction Quassinoids are natural products formed through the oxidative degradation of triterpene derivatives with anti-inflammatory, antimicrobial, antineoplastic, and antiplasmodial effects (Chakraborty and Pal, 2013; Houël et al., 2013). They are characteristic ingredients of the family Simar
-
Based on the above we hypothesized that a PROTAC strategy
2020-03-24
Based on the above, we hypothesized that a PROTAC strategy would be effective to induce CK2 protein degradation. Herein reported is an approach for the preparation of novel PROTACs via “click reaction” for degradation of CK2 protein (Fig. 2). Importantly, “click reaction” is a very facile, selective
-
Concerning family A GPCRs although it has been described
2020-03-23
Concerning family A GPCRs, although it has been described that several receptors are able to operate as monomers (Arcemisbéhère et al., 2010; Bayburt et al., 2011; Chabre & le Maire, 2005; Ernst, Gramse, Kolbe, Hofmann, & Heck, 2007; Hanson et al., 2007; Kuszak et al., 2009; Whorton et al., 2007), e
-
Studies of LRRK tagged with green fluorescent protein
2020-03-23
Studies of LRRK2 tagged with green fluorescent protein revealed the presence of monomeric LRRK2 in the PD128907 HCl australia of living cells, while LRRK2 oligomers were found in the proximity of the plasma membrane, suggesting that both oligomerization status and compartmentalization of LRRK2 defi
-
Darifenacin HBr receptor A genomic DNA sequence was
2020-03-23
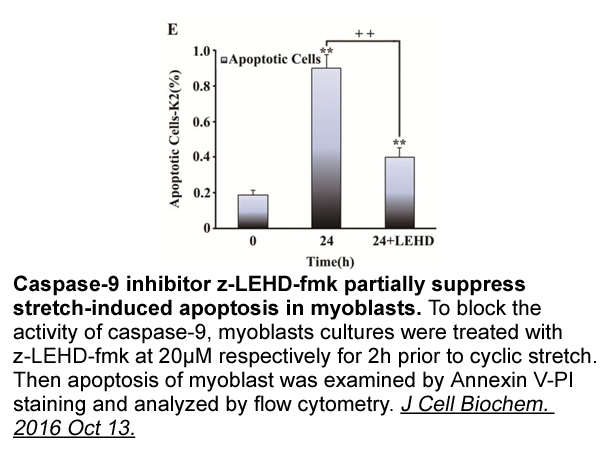
A genomic DNA sequence was retrieved (Scaffold_30) from the fugu genomic database ( v3.0), which encodes the homolog of CXCL8 of many other vertebrates and specific primers CXCL8F1 and CXCL8R1 () were designed for 3′-RACE and 5′-RACE PCR, respectively. First-strand thymus cDNA was used as template.
-
In the preclinical evaluation of CRF receptor antagonists ef
2020-03-23
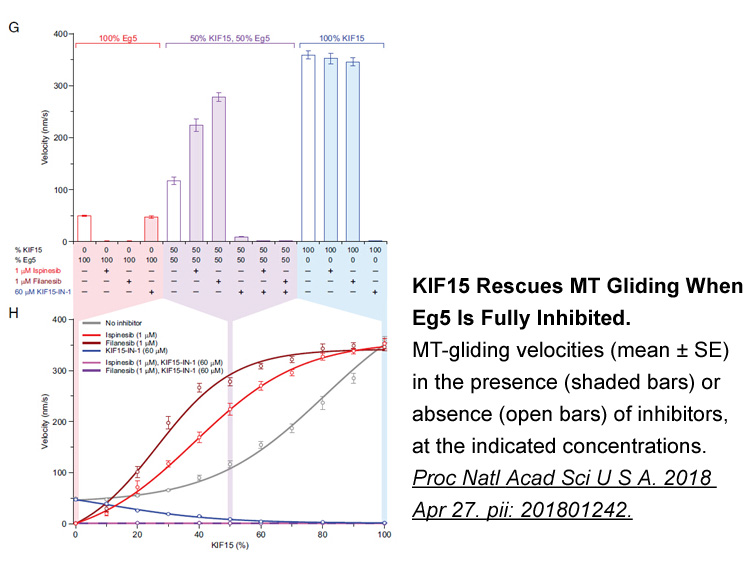
In the preclinical evaluation of CRF1 receptor antagonists, effects were mainly focused on the regulation of hyperactivated HPA axis and the modulation of anxiety or depressive-like behaviors in CRF challenge [16], [17], [18] or stress models [16], [19], [20]. However, it remains to be concluded whe
-
The inhibition of FAS by C produces an
2020-03-23
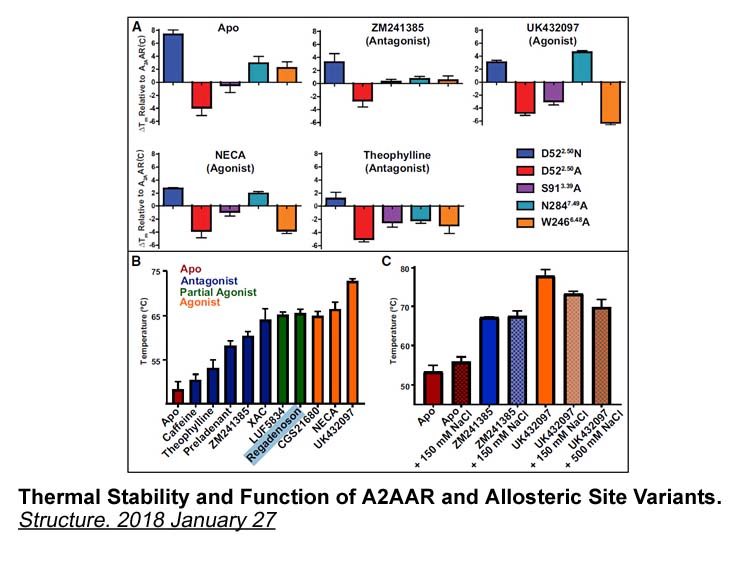
The inhibition of FAS by C75 produces an accumulation of malonyl-CoA which is difficult to reconcile with the activation of CPT1 reported by others [2], [16], [17], [18]. To unravel this paradox the mechanism of action of C75 needs to be examined. We recently demonstrated that C75 is converted in vi
-
Other membrane currents are affected as well Some
2020-03-23
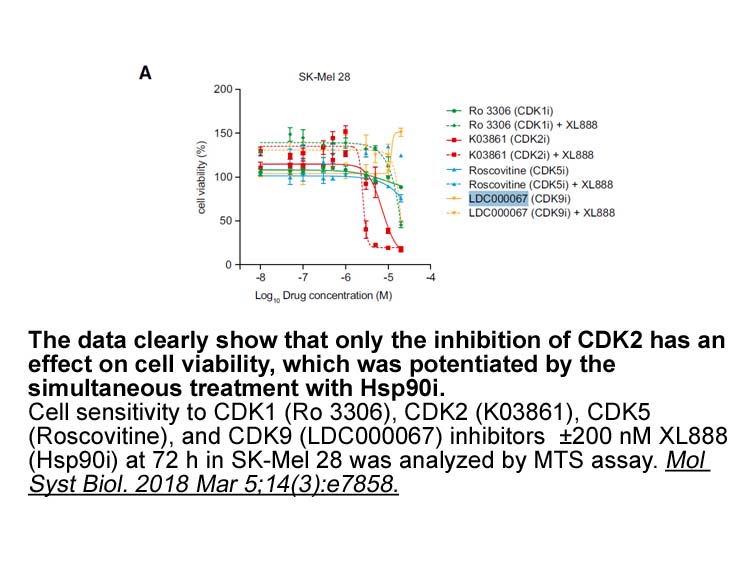
Other membrane currents are affected as well. Some studies have provided evidence that sulfonylureas, in addition to blocking KATP channels, also inhibit chloride and calcium channels. GLYB has been shown to almost inhibit the current generated by Na+–K+ pumps in a concentration-dependent manner (wi
-
Early tubular differentiation in WT lacks lumen formation
2020-03-23

Early tubular differentiation in WT lacks lumen formation and, therefore, sometimes can be confused with rosettes of ES/PNET or neuroblastoma. However, tubular structure usually has HO-3867 neatly aligned to form a single layer around the future lumen, whereas neuroblastic rosettes are arranged more
-
Real time PCR as described previously with minor
2020-03-23

Real-time PCR, as described previously with minor modifications, was used to detect DNA . The PCR mixture (Qiagen) was as follows: 5mM MgCl2, 2.5μl of 10x PCR Buffer, 0.125μl of HotStarTaq DNA Polymerase, 0.25mM of each dNTP, 0.4μM of each primer and 0.1μM of FAMlabelled probe (‘5-aacaattggagggcaag
16091 records 866/1073 page Previous Next First page 上5页 866867868869870 下5页 Last page